Conifers are ecologically and economically most important gymnosperms dominating many terrestrial ecosystems in the Northern Hemisphere. Fossil records show the presence of gymnosperms in the Late Carboniferous period 300Mya. They thrived through the age of the dinosaurs before and after mass extinction events 250 – 65 Mya. Conifers are represented by 630 species belonging to eight families and 70 genera. Some of the woody plant species are the longest living and largest living organisms on this planet. The enormous size of their genome 20 – 40 giga base pairs which is seven times the human genome was a major challenge in conifer genomics. The advancements in sequencing and bioinformatics technology have made the genome analyses possible today. The draft assemblies are providing valuable insight into the genome organisation, gene families involved in metabolic pathways, genetic basis of environment – plant interaction, comparative and evolutionary analysis of angio- and gymnosperms. They are also going to provide tools for better forest management strategies and markers for genome selection for tree breeding and improvement for well adapted forests.

© Gerhard Leubner 2005″ and cite the source “The Seed Biology Place – http://www.seedbiology.de/”
This paper prepared a context for projects like ProCoGen highlighting the unique features of conifers known until recently. It emphasises the importance of conifer genome sequencing projects in advancing the fundamental understanding of gene regulation, especially genes involved in abiotic and biotic stress response, disease resistance and wood formation. Identification of markers for genome selection methods of tree breeding for well adapted trees by exploiting ecological and economic value contained in the genetic information.
Recent advancements in technology have made sequencing of enormous conifer genomes feasible. Based on a comparative study of recently published sequence data from Picea glauca, P. abies and Pinus taeda the authors have tried to bring distinctive features of the conifer genomes to light especially composition and structure and how they differ from angiosperms, using Amborella genome as a key reference for this comparison. The protein coding sequences are not relatively larger than angiosperms. Apart from one ancient genome duplication event dating back to the angio-gymnosperm spilt ca. 350Mya there has be no evidence of such an event in shaping gymnosperm genomes unlike the angiosperms where several whole genome and smaller duplication events have shaped genomes, diversification and evolution. The conifer genomes owe their size to accumulation of transposable elements mostly LTR-RTs (long terminal repeat retrotransposons), and some LINEs (long interspersed nuclear elements), SINEs (short interspersed nuclear elements) and DNA transposons. Nystedt et al. 2013 also found similar evidence in his comparative study of gymnosperm genomes.
Long introns were also abundant in the three conifer genomes. Small non coding RNA and short RNA (sRNAs) were also different between angio- and gymnosperms. 21 nucleotide sRNA were more abundant than 24 nucleotide sRNAs in conifers indicating difference in epigenetic response and gene regulation in conifers as compared to angiosperms. Genetic bases of difference between angio- and gymnosperm reproductive systems, water conducting vessel, wood formation, and secondary metabolism could be traced back by phylogenetic analysis following the evolutionary trajectory of gene families involved. Authors review findings of comparative genomics that reveal conifer/gymnosperm specific gene families and their evolution. They also review the findings about genes involved in wood formation, abiotic and biotic stress response, somatic embryogenesis, disease resistance. Development of high-throughput genotyping chips is being accelerated by the availability of transcriptomic data. This would aid the genome wide association studies and tree breeding.
Suggested reading:
 An introduction to next generation sequencing: from Illumina
An introduction to next generation sequencing: from Illumina
This paper gives an overview of genome sequencing in non-model species, its applications and implications for future research.
Krutovsky K. et al. 2004. Comparative mapping in Pinaceae. Genetics 168: 447 – 461.
Revealed extensive synteny and colinearity between pine and spruce, implying that clusters of gene functions are preserved
Applying the next generation sequencing technology the authors have been able to assemble the transcribed regions of the maritime pine genome. The study provides an inventory of expressed genes and an exhaustive characterisation of the protein coding regions. It throws light on the gene family constitution, genes involved in metabolic pathways and the transcription factors composition for understanding gene regulatory networks. The sequencing information also facilitated the identification of polymorphism and establishment of single nucleotide polymorphism (SNP) and simple sequence repeat (SSR) database for genotyping application and translational genomic information relevant for tree improvement programmes. The research outcomes would also aid in future genome assembly, functional genomics, evolutionary and comparative studies as well as applied studies of maritime pines.
Polymorphism detection and gene mapping using exome capture has an advantage that it helps detect segregating markers in a population.
The authors have published the most saturated gene based map in conifers. Markers are like landmarks that can aid in locating position of genes on chromosomes. This gene based map would be helpful in constructing a pine reference sequence as well as other applied research. “Because of interspecific synteny the gene based maps would be useful to support genome assembly and analysis”
The loblolly pine v1.01 assembly has the largest number of annotations among the conifer reference genomes that have been sequenced so far. This draft genome assembly reveals features of conifer genome organisation, aids comparative analyses & evolution of angiosperms and gymnosperms, and identification of genes regulating ecological responses and controlling economically important traits like wood formation. An account of the conifer genome evolution is also given by Nystedt B. 2013 Nature 497: 579–584 doi:10.1038/nature12211. Yet another genome assembly of white spruce using whole genome shotgun sequencing provides valuable information to support forest management strategies and understand tree and environmental interactions at gene level. Birol et al. 2013 Bioinformatics doi: 10.1093/bioinformatics/btt178
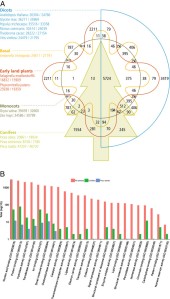 Fig (A) Identification of orthologous groups of genes for 14 species split into five categories: conifers (Picea abies, Picea sitchensis, and Pinus taeda), monocots (Oryza sativa and Zea mays), dicots (Arabidopsis thaliana, Glycine max, Populus trichocarpa, Ricinus communis, Theobroma cacao, and Vitis vinifera), early land plants (Selaginella moellendorffii and Physcomitrella patens), and a basal angiosperm (Amborella trichopoda). Here, we depict the number of clusters in common between the biological categories in the intersections. The total number of sequences for each species is provided under the name (total number of sequences/total number of clustered sequences). (B) Gene ontology molecular function term assignments by family for all species (red), conifers (green), and Pinus taeda exclusively (blue).
Fig (A) Identification of orthologous groups of genes for 14 species split into five categories: conifers (Picea abies, Picea sitchensis, and Pinus taeda), monocots (Oryza sativa and Zea mays), dicots (Arabidopsis thaliana, Glycine max, Populus trichocarpa, Ricinus communis, Theobroma cacao, and Vitis vinifera), early land plants (Selaginella moellendorffii and Physcomitrella patens), and a basal angiosperm (Amborella trichopoda). Here, we depict the number of clusters in common between the biological categories in the intersections. The total number of sequences for each species is provided under the name (total number of sequences/total number of clustered sequences). (B) Gene ontology molecular function term assignments by family for all species (red), conifers (green), and Pinus taeda exclusively (blue).
20.15 billion bp draft genome whole genome shot gun sequencing . Most complete and contiguous genome sequence available for conifers.
Wegrzyn J. L. et al. 2014. Unique features of the loblolly pine (Pinus taeda) megagenome revealed through sequence annotation. Genetics 196: 891-909.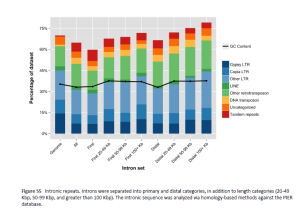
As the title suggests this paper provides information derived from sequencing and annotation of the loblolly pine genome especially the organisation of the genome, its size, content and structure. These findings will be a great asset in comparative genomics and evolutionary studies involving gymnosperms.
Sena J. S. et al. 2014. Evolution of gene structure in the conifer Picea glauca: comparative analysis of the impact of intron size. BMC Plant Biology 14: 95
Gene structure of 35 genes of Picea glauca was analysed and also compared to sequenced genomes of angiosperms. Intron size and distribution varied according to the expression profiles. The position of long introns within genes was highly conserved in conifers. LTRs had higher repetitive element representation in introns. Copia and gypsy were longer in exons that in introns.
Conserved gene structure, intron size, large no. of repetitive elements, small gene space were products of complex evolutionary history of conifers.
